Orthology Basics
The study of genetic material almost always starts with identifying, within or across species, homologous regions—regions of common ancestry. It is useful to distinguish between two classes of homologous genes: orthologs, which are pairs of genes that started diverging via evolutionary speciation, and paralogs, which are pairs of genes that started diverging via gene duplication.
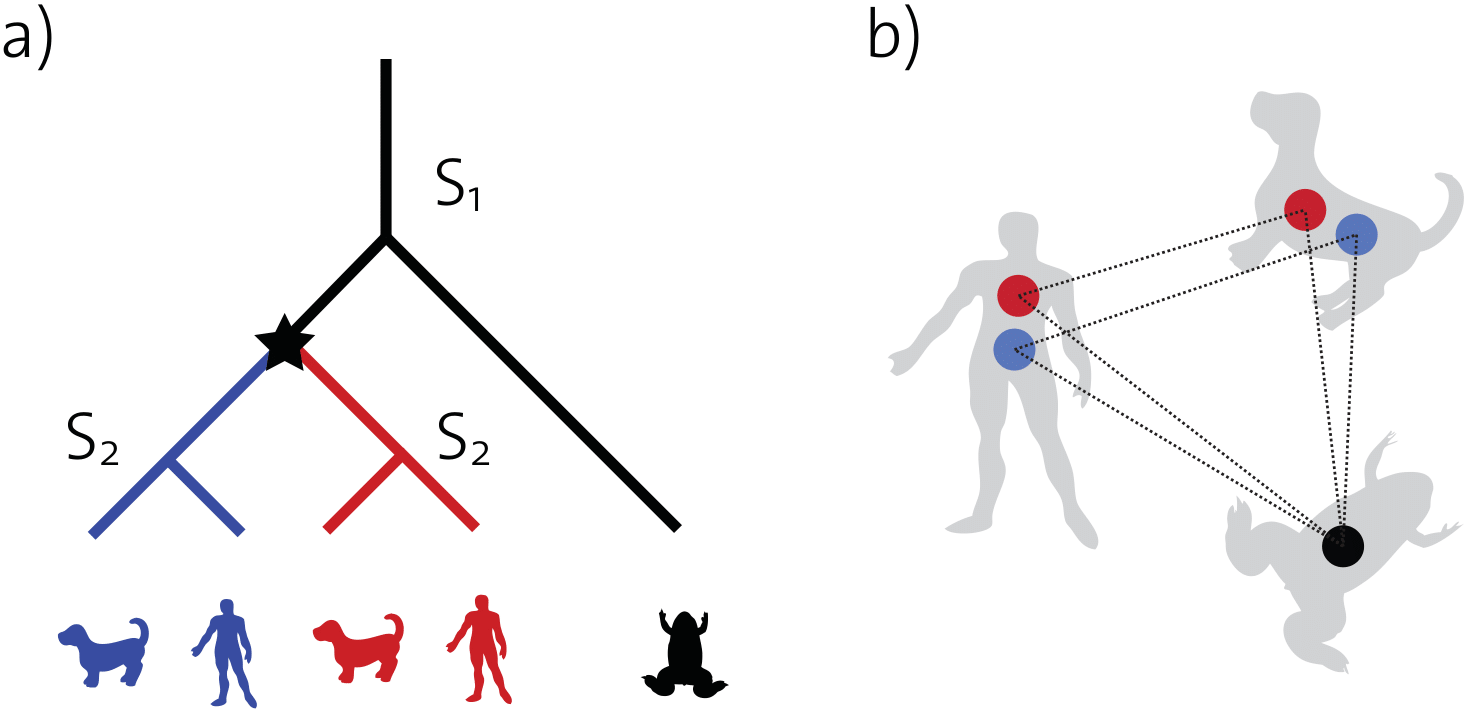
(b) The corresponding orthology graph. The genes are represented here by vertices and orthology relationships by edges. The frog gene forms one-to-many orthology with both the human and dog genes, because it is orthologous to more than one sequence in each of these organisms.
In comparative genomics and phylogenetics, the fundamental concept of orthology relates “corresponding” genes in different species: orthologs are pairs of genes which have evolved from a single gene in the last common ancestor. Among many applications, orthologs are useful to infer species trees and tend to be functionally conserved.
Please see (Altenhoff et al. 2019), (Zahn-Zabal et al. 2020) for more information.
Orthology Definitions
Orthology is a relation defined over a pair of homologous genes, where the two genes have emerged through a speciation event.
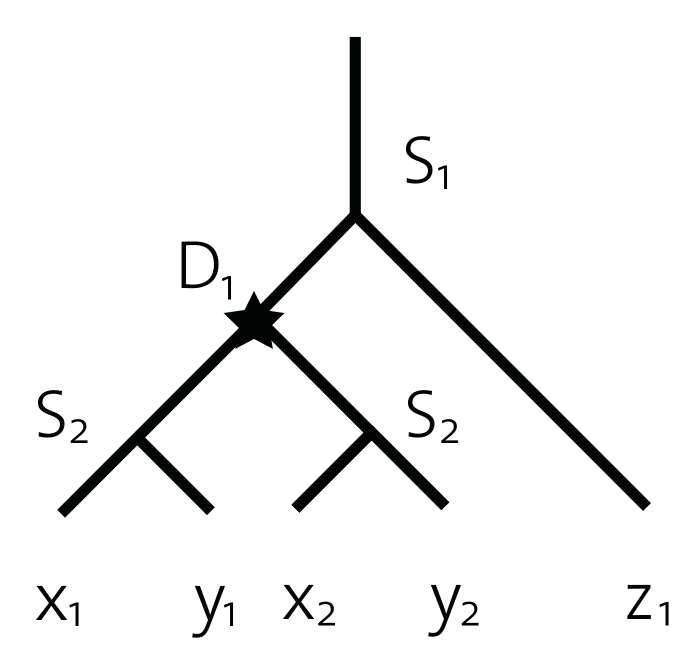
Example pairs of orthologs are (x1, y1) or (x2, z1). Orthologs can be further subclassified into one-to-one, one-to-many, many-to-one, and many-to-many orthologs. The qualifiers one and many indicate for each of the two involved genes whether they underwent an additional duplication after the speciation between the two genomes. Hence, the gene pair (x1, y1) is an example of a one-to-one orthologous pair, whereas (x2, z1) is a many-to-one ortholog relation.
Paralogy is a relation defined over a pair of homologous genes that have emerged through a gene duplication, e.g., (x1, x2) or (x1, y2).
In-Paralogy is a relation defined over a triplet. It involves a pair of genes and a speciation event of reference. A gene pair is an in-paralog if they are paralogs and duplicated after the speciation event of reference. The pair (x1, y2) are in-paralogs with respect to the speciation event S1.
Out-Paralogy is also a relation defined over a pair of genes and a speciation event of reference. This pair is out-paralogs if the duplication event through which they are related to each other predates the speciation event of reference. Hence, the pair (x1, y2) are out-paralogs with respect to the speciation event S2.
Co-orthology is a relation defined over three genes, where two of them are in-paralogs with respect to the speciation event associated with the third gene. The two in-paralogous genes are said to be co-orthologous to the third (out-group) gene. Thus, x1 and y2 are co-orthologs with respect to z1.
Homoeology is a specific type of homologous relation in a polyploid species, which thus contain multiple “sub-genomes.” This relation describes pairs of genes that originated by speciation and were brought back together in the same genome by allopolyploidization (hybridization). Thus, in the absence of rearrangement, homoeologs can be thought of as orthologs between sub-genomes.
Please see (Altenhoff et al. 2019), (Glover et al. 2016) for more information.